Run the MCMC algorithm for a BART model for count-valued responses using STAR. The transformation can be known (e.g., log or sqrt) or unknown (Box-Cox or estimated nonparametrically) for greater flexibility.
Usage
bart_star(
y,
X,
X_test = NULL,
y_test = NULL,
transformation = "np",
y_max = Inf,
n.trees = 200,
sigest = NULL,
sigdf = 3,
sigquant = 0.9,
k = 2,
power = 2,
base = 0.95,
nsave = 1000,
nburn = 1000,
nskip = 0,
save_y_hat = FALSE,
verbose = TRUE
)Arguments
- y
n x 1vector of observed counts- X
n x pmatrix of predictors- X_test
n0 x pmatrix of predictors for test data- y_test
n0 x 1vector of the test data responses (used for computing log-predictive scores)- transformation
transformation to use for the latent process; must be one of
"identity" (identity transformation)
"log" (log transformation)
"sqrt" (square root transformation)
"np" (nonparametric transformation estimated from empirical CDF)
"pois" (transformation for moment-matched marginal Poisson CDF)
"neg-bin" (transformation for moment-matched marginal Negative Binomial CDF)
"box-cox" (box-cox transformation with learned parameter)
"ispline" (transformation is modeled as unknown, monotone function using I-splines)
- y_max
a fixed and known upper bound for all observations; default is
Inf- n.trees
number of trees to use in BART; default is 200
- sigest
positive numeric estimate of the residual standard deviation (see ?bart)
- sigdf
degrees of freedom for error variance prior (see ?bart)
- sigquant
quantile of the error variance prior that the rough estimate (sigest) is placed at. The closer the quantile is to 1, the more aggressive the fit will be (see ?bart)
- k
the number of prior standard deviations E(Y|x) = f(x) is away from +/- 0.5. The response is internally scaled to range from -0.5 to 0.5. The bigger k is, the more conservative the fitting will be (see ?bart)
- power
power parameter for tree prior (see ?bart)
- base
base parameter for tree prior (see ?bart)
- nsave
number of MCMC iterations to save
- nburn
number of MCMC iterations to discard
- nskip
number of MCMC iterations to skip between saving iterations, i.e., save every (nskip + 1)th draw
- save_y_hat
logical; if TRUE, compute and save the posterior draws of the expected counts, E(y), which may be slow to compute
- verbose
logical; if TRUE, print time remaining
Value
a list with the following elements:
post.pred: draws from the posterior predictive distribution ofypost.sigma: draws from the posterior distribution ofsigmapost.log.like.point: draws of the log-likelihood for each of thenobservationsWAIC: Widely-Applicable/Watanabe-Akaike Information Criterionp_waic: Effective number of parameters based on WAICpost.pred.test: draws from the posterior predictive distribution at the test pointsX_test(NULLifX_testis not given)post.fitted.values.test: posterior draws of the conditional mean at the test pointsX_test(NULLifX_testis not given)post.mu.test: draws of the conditional mean of z_star at the test pointsX_test(NULLifX_testis not given)post.log.pred.test: draws of the log-predictive distribution for each of then0test cases (NULLifX_testis not given)fitted.values: the posterior mean of the conditional expectation of the countsy(NULLifsave_y_hat=FALSE)post.fitted.values: posterior draws of the conditional mean of the countsy(NULLifsave_y_hat=FALSE)
In the case of transformation="ispline", the list also contains
post.g: draws from the posterior distribution of the transformationgpost.sigma.gamma: draws from the posterior distribution ofsigma.gamma, the prior standard deviation of the transformation g() coefficients
If transformation="box-cox", then the list also contains
post.lambda: draws from the posterior distribution oflambda
Details
STAR defines a count-valued probability model by (1) specifying a Gaussian model for continuous *latent* data and (2) connecting the latent data to the observed data via a *transformation and rounding* operation. Here, the model in (1) is a Bayesian additive regression tree (BART) model.
Posterior and predictive inference is obtained via a Gibbs sampler that combines (i) a latent data augmentation step (like in probit regression) and (ii) an existing sampler for a continuous data model.
There are several options for the transformation. First, the transformation
can belong to the *Box-Cox* family, which includes the known transformations
'identity', 'log', and 'sqrt', as well as a version in which the Box-Cox parameter
is inferred within the MCMC sampler ('box-cox'). Second, the transformation
can be estimated (before model fitting) using the empirical distribution of the
data y. Options in this case include the empirical cumulative
distribution function (CDF), which is fully nonparametric ('np'), or the parametric
alternatives based on Poisson ('pois') or Negative-Binomial ('neg-bin')
distributions. For the parametric distributions, the parameters of the distribution
are estimated using moments (means and variances) of y. Third, the transformation can be
modeled as an unknown, monotone function using I-splines ('ispline'). The
Robust Adaptive Metropolis (RAM) sampler is used for drawing the parameter
of the transformation function.
Examples
# \donttest{
# Simulate data with count-valued response y:
sim_dat = simulate_nb_friedman(n = 100, p = 5)
y = sim_dat$y; X = sim_dat$X
# BART-STAR with log-transformation:
fit_log = bart_star(y = y, X = X, transformation = 'log',
save_y_hat = TRUE, nburn=1000, nskip=0)
#> [1] "Burn-In Period"
#> [1] "Starting sampling"
#> [1] "0 seconds remaining"
#> [1] "Total time: 2 seconds"
# Fitted values
plot_fitted(y = sim_dat$Ey,
post_y = fit_log$post.fitted.values,
main = 'Fitted Values: BART-STAR-log')
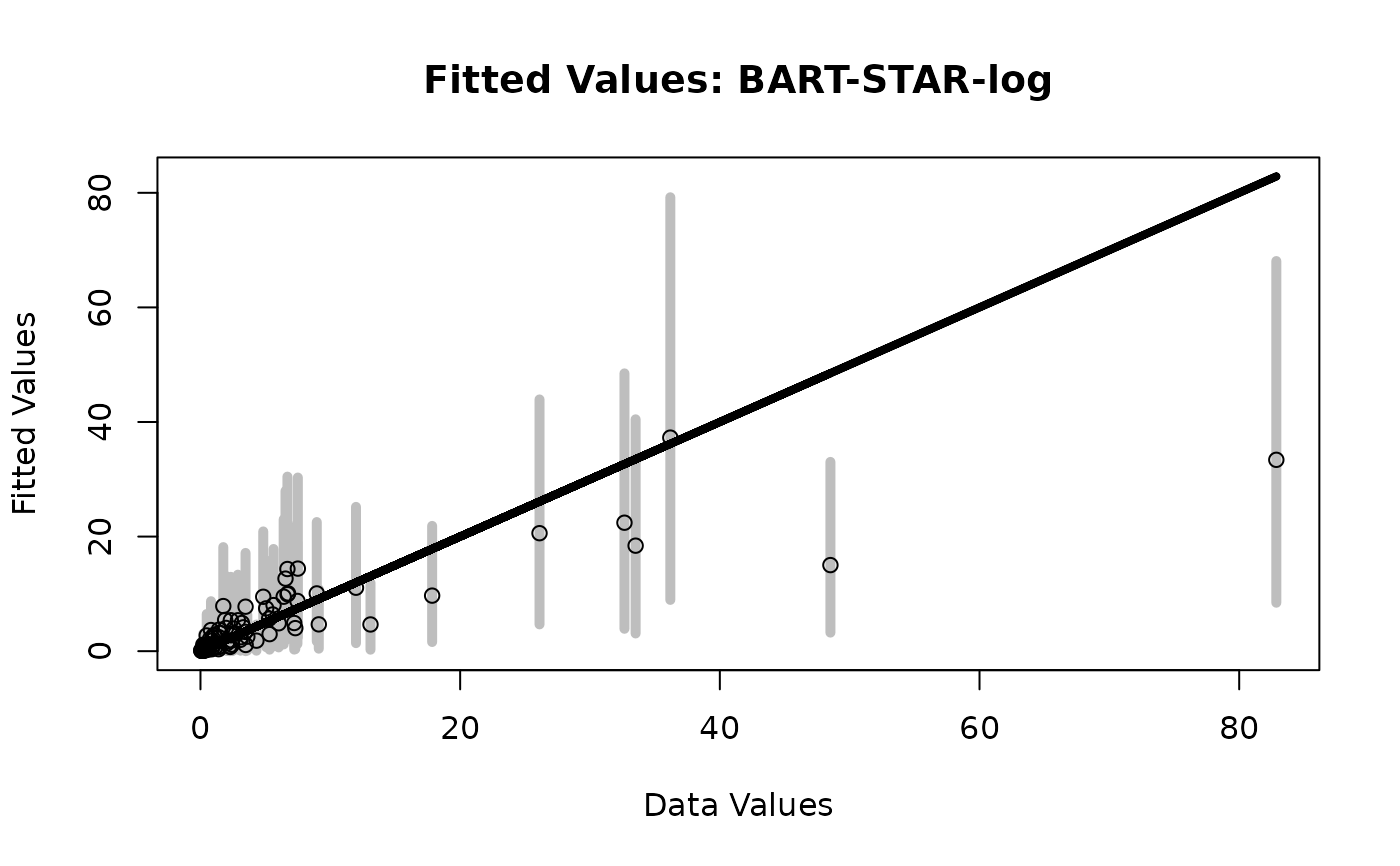 # WAIC for BART-STAR-log:
fit_log$WAIC
#> [1] 382.4023
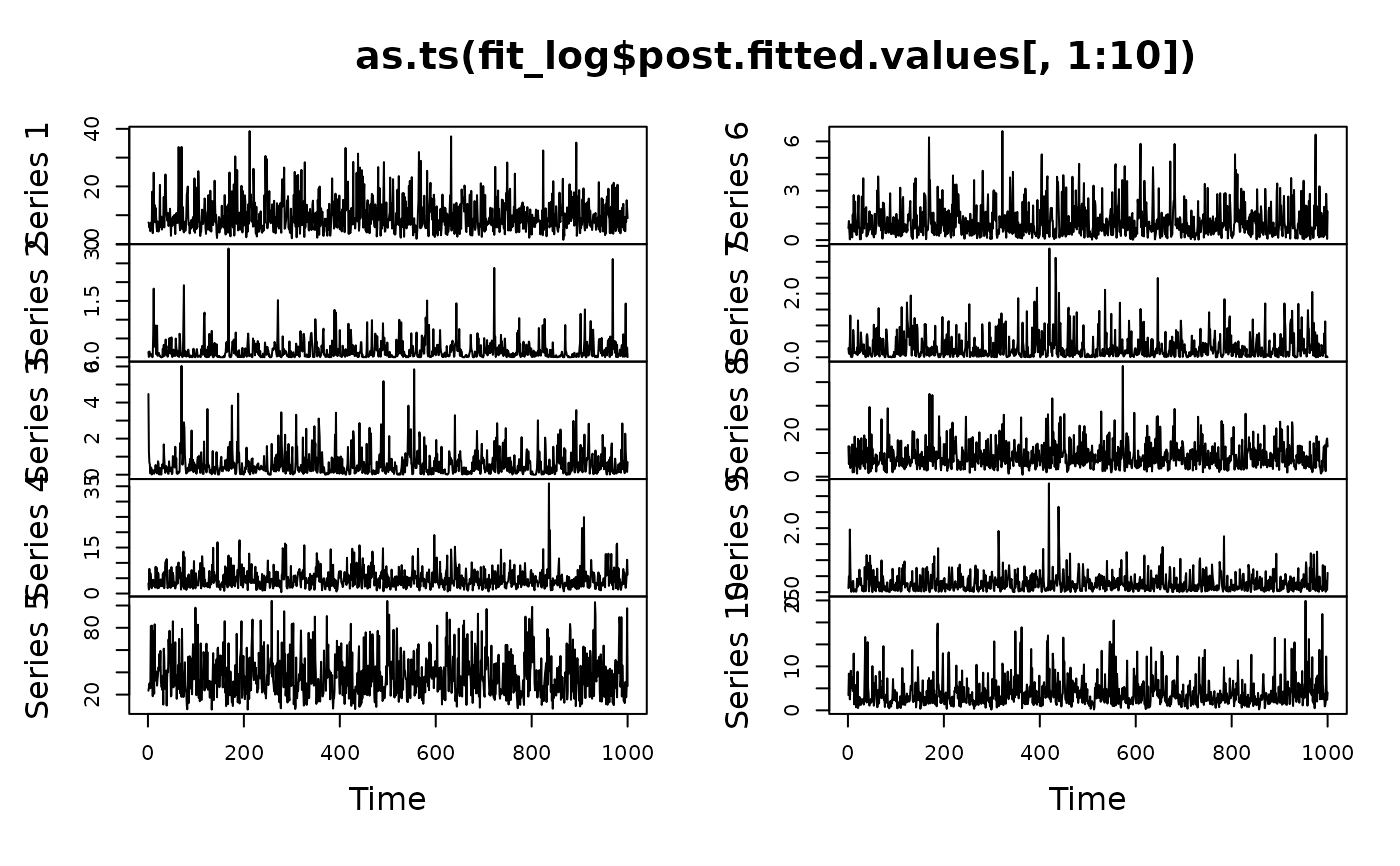
# MCMC diagnostics:
plot(as.ts(fit_log$post.fitted.values[,1:10]))
# WAIC for BART-STAR-log:
fit_log$WAIC
#> [1] 382.4023
# MCMC diagnostics:
plot(as.ts(fit_log$post.fitted.values[,1:10]))
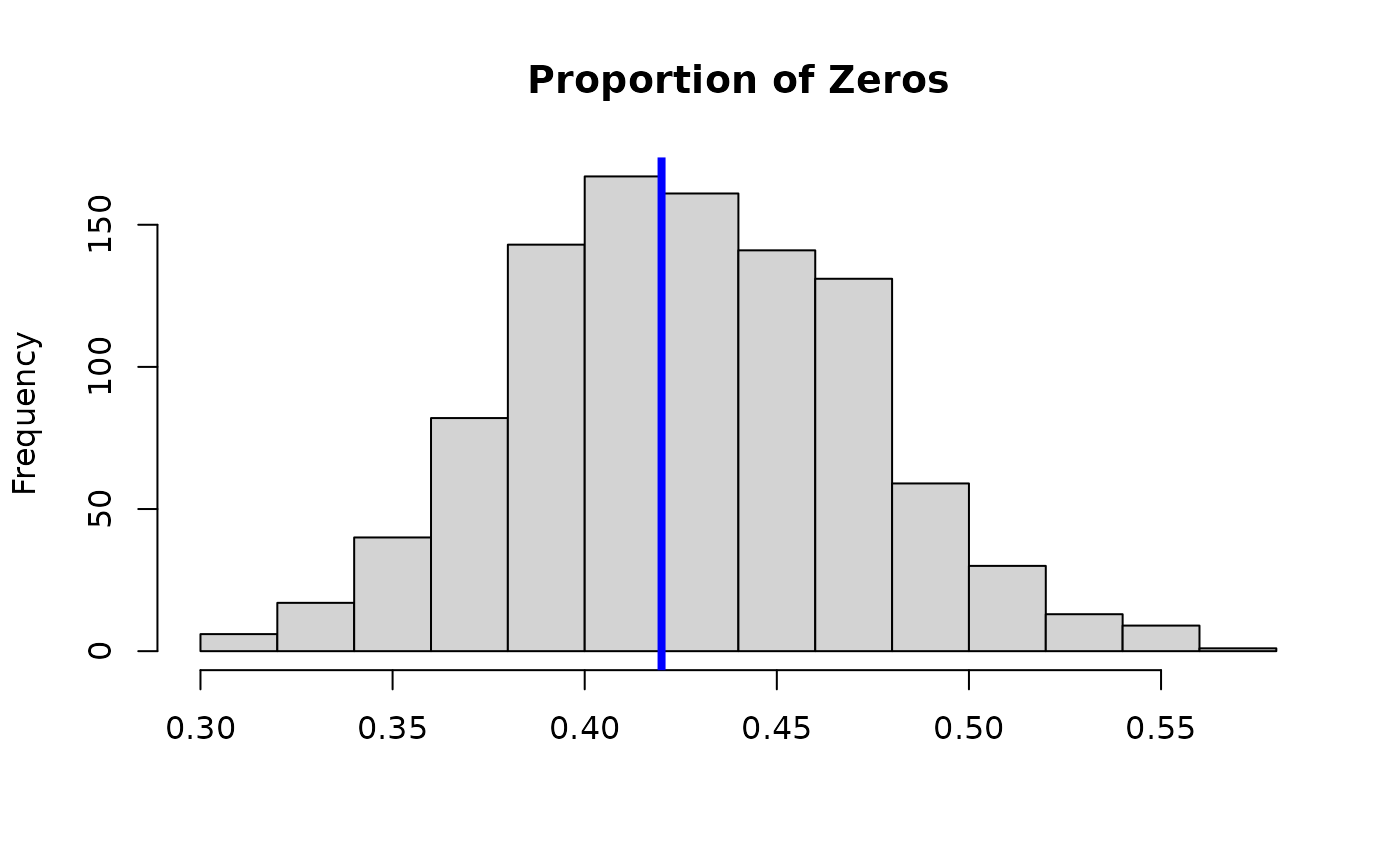
 # Posterior predictive check:
hist(apply(fit_log$post.pred, 1,
function(x) mean(x==0)), main = 'Proportion of Zeros', xlab='');
abline(v = mean(y==0), lwd=4, col ='blue')
# Posterior predictive check:
hist(apply(fit_log$post.pred, 1,
function(x) mean(x==0)), main = 'Proportion of Zeros', xlab='');
abline(v = mean(y==0), lwd=4, col ='blue')
 # BART-STAR with nonparametric transformation:
fit = bart_star(y = y, X = X,
transformation = 'np', save_y_hat = TRUE)
#> [1] "Burn-In Period"
#> [1] "Starting sampling"
#> [1] "0 seconds remaining"
#> [1] "Total time: 4 seconds"
# Fitted values
plot_fitted(y = sim_dat$Ey,
post_y = fit$post.fitted.values,
main = 'Fitted Values: BART-STAR-np')
# BART-STAR with nonparametric transformation:
fit = bart_star(y = y, X = X,
transformation = 'np', save_y_hat = TRUE)
#> [1] "Burn-In Period"
#> [1] "Starting sampling"
#> [1] "0 seconds remaining"
#> [1] "Total time: 4 seconds"
# Fitted values
plot_fitted(y = sim_dat$Ey,
post_y = fit$post.fitted.values,
main = 'Fitted Values: BART-STAR-np')
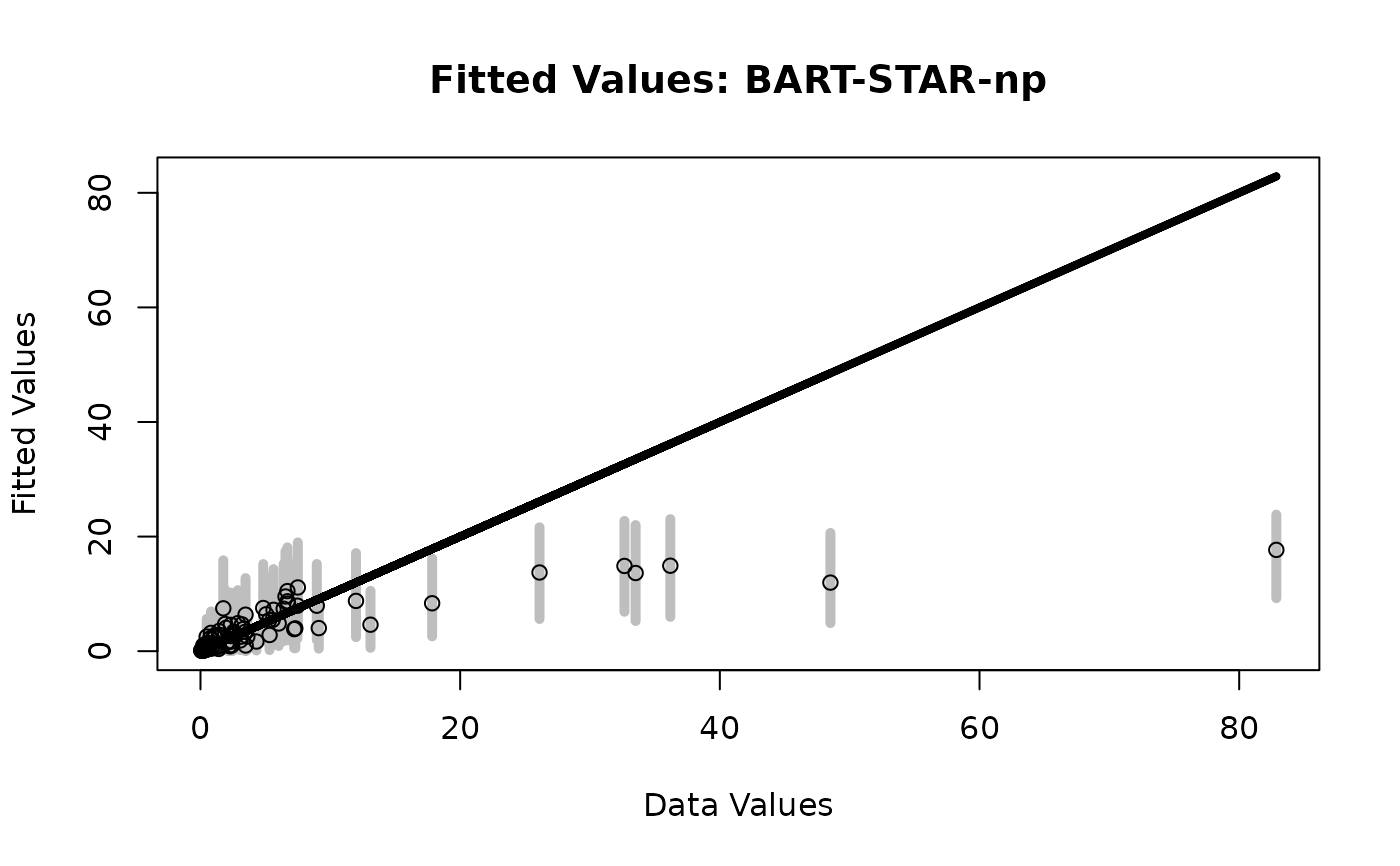 # WAIC for BART-STAR-np:
fit$WAIC
#> [1] 379.1259
# MCMC diagnostics:
plot(as.ts(fit$post.fitted.values[,1:10]))
# WAIC for BART-STAR-np:
fit$WAIC
#> [1] 379.1259
# MCMC diagnostics:
plot(as.ts(fit$post.fitted.values[,1:10]))
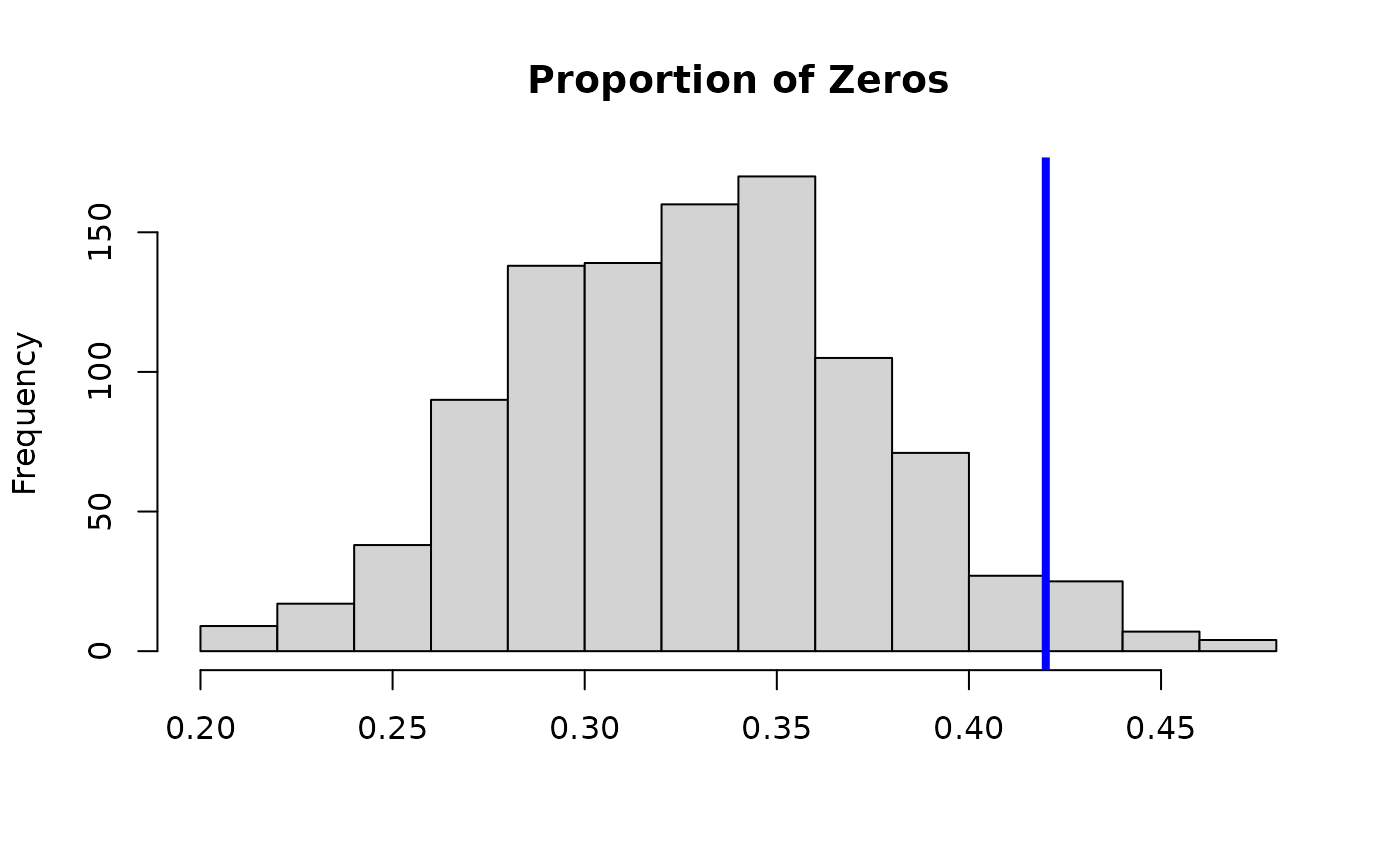
 # Posterior predictive check:
hist(apply(fit$post.pred, 1,
function(x) mean(x==0)), main = 'Proportion of Zeros', xlab='');
abline(v = mean(y==0), lwd=4, col ='blue')
# Posterior predictive check:
hist(apply(fit$post.pred, 1,
function(x) mean(x==0)), main = 'Proportion of Zeros', xlab='');
abline(v = mean(y==0), lwd=4, col ='blue')
 # }
# }