Compute samples from the predictive distributions of a STAR spline regression model using either a Gibbs sampling approach or exact Monte Carlo sampling (default is Gibbs sampling which scales better for large n).
Usage
spline_star(
y,
tau = NULL,
transformation = "np",
y_max = Inf,
psi = NULL,
nsave = 1000,
use_MCMC = TRUE,
nburn = 1000,
nskip = 0,
verbose = TRUE
)Arguments
- y
n x 1vector of observed counts- tau
n x 1vector of observation points; if NULL, assume equally-spaced on [0,1]- transformation
transformation to use for the latent data; must be one of
"identity" (identity transformation)
"log" (log transformation)
"sqrt" (square root transformation)
"bnp" (Bayesian nonparametric transformation using the Bayesian bootstrap)
"np" (nonparametric transformation estimated from empirical CDF)
"pois" (transformation for moment-matched marginal Poisson CDF)
"neg-bin" (transformation for moment-matched marginal Negative Binomial CDF)
- y_max
a fixed and known upper bound for all observations; default is
Inf- psi
prior variance (1/smoothing parameter); if NULL, update in MCMC
- nsave
number of MCMC iterations to save (or number of Monte Carlo simulations)
- use_MCMC
logical; whether to run Gibbs sampler or Monte Carlo (default is TRUE)
- nburn
number of MCMC iterations to discard
- nskip
number of MCMC iterations to skip between saving iterations, i.e., save every (nskip + 1)th draw
- verbose
logical; if TRUE, print time remaining
Value
a list with the following elements:
post.pred:nsave x nsamples from the posterior predictive distribution at the observation pointstaumarg_like: the marginal likelihood (only ifuse_MCMC=FALSE; otherwise NULL)
Details
STAR defines a count-valued probability model by (1) specifying a Gaussian model for continuous *latent* data and (2) connecting the latent data to the observed data via a *transformation and rounding* operation. Here, the continuous latent data model is a spline regression.
There are several options for the transformation. First, the transformation
can belong to the *Box-Cox* family, which includes the known transformations
'identity', 'log', and 'sqrt'. Second, the transformation
can be estimated (before model fitting) using the empirical distribution of the
data y. Options in this case include the empirical cumulative
distribution function (CDF), which is fully nonparametric ('np'), or the parametric
alternatives based on Poisson ('pois') or Negative-Binomial ('neg-bin')
distributions. For the parametric distributions, the parameters of the distribution
are estimated using moments (means and variances) of y. The distribution-based
transformations approximately preserve the mean and variance of the count data y
on the latent data scale, which lends interpretability to the model parameters.
Lastly, the transformation can be modeled using the Bayesian bootstrap ('bnp'),
which is a Bayesian nonparametric model and incorporates the uncertainty
about the transformation into posterior and predictive inference.
Note
For the 'bnp' transformation
there are numerical stability issues when psi is modeled as unknown.
In this case, it is better to fix psi at some positive number.
Examples
# Simulate some data:
n = 100
tau = seq(0,1, length.out = n)
y = round_floor(exp(1 + rnorm(n)/4 + poly(tau, 4)%*%rnorm(n=4, sd = 4:1)))
# Sample from the predictive distribution of a STAR spline model:
fit = spline_star(y = y, tau = tau)
#> [1] "Burn-In Period"
#> [1] "Starting sampling"
#> [1] "0 seconds remaining"
#> [1] "Total time: 1 seconds"
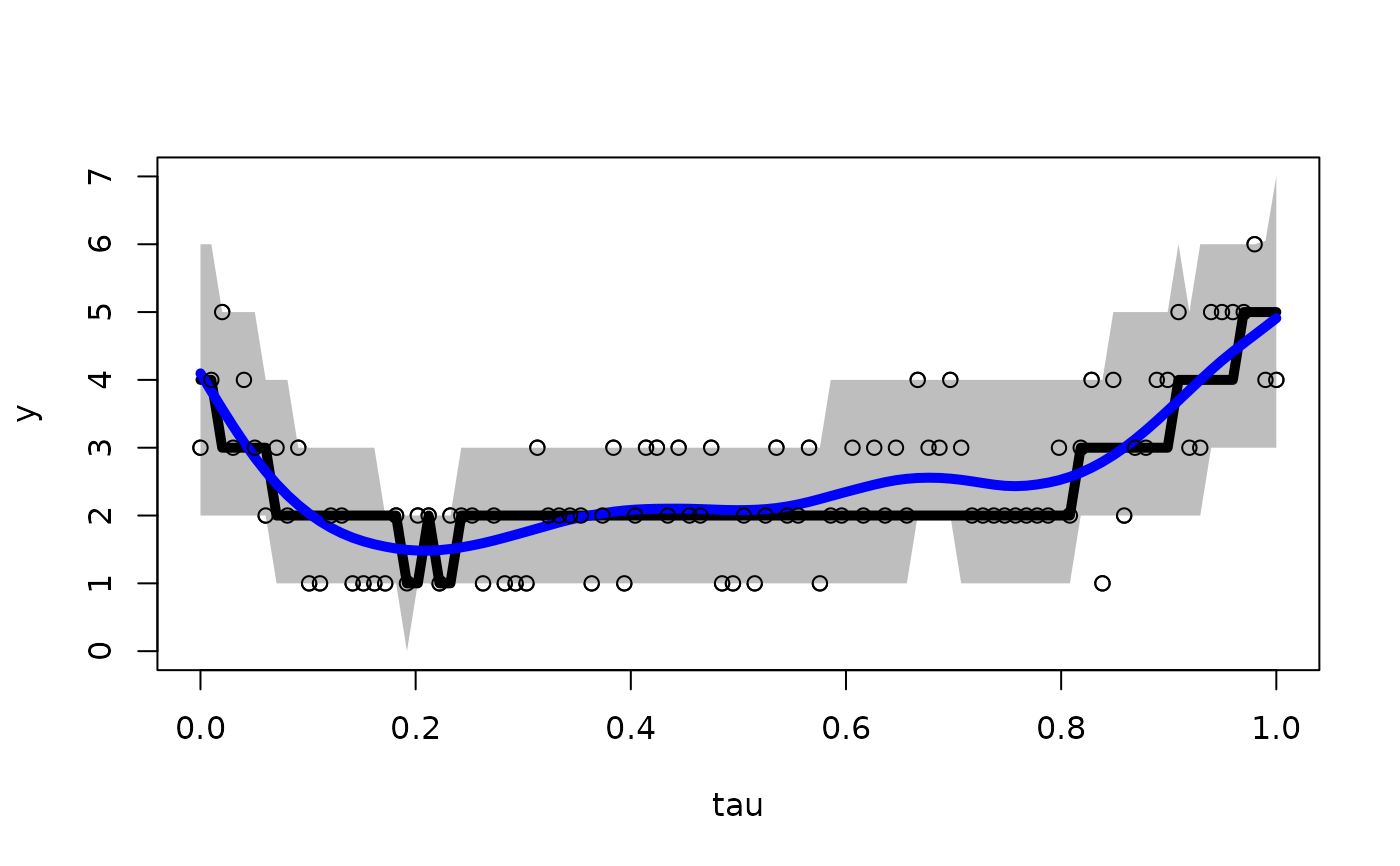
# Compute 90% prediction intervals:
pi_y = t(apply(fit$post.pred, 2, quantile, c(0.05, .95)))
# Plot the results: intervals, median, and smoothed mean
plot(tau, y, ylim = range(pi_y, y))
polygon(c(tau, rev(tau)),c(pi_y[,2], rev(pi_y[,1])),col='gray', border=NA)
lines(tau, apply(fit$post.pred, 2, median), lwd=5, col ='black')
lines(tau, smooth.spline(tau, apply(fit$post.pred, 2, mean))$y, lwd=5, col='blue')
lines(tau, y, type='p')